|
サイズ: 29795
編集者: intergroup01
コメント:
|
サイズ: 29798
編集者: intergroup01
コメント:
|
| 削除された箇所はこのように表示されます。 | 追加された箇所はこのように表示されます。 |
| 行 75: | 行 75: |
| http://imc.tut.ac.jp/research/form. | http://imc.tut.ac.jp/en/research/form. |
Using Cluster Systems
This section explains the usage methods common among the Next Generation Simulation Technology Education and the Wide-Area Coordinated Cluster System for Education and Research. See the page for each system for details of specific functions.
目次
Logging In
* Computer Systems for the Next Generation Simulation Technology Education*/
To compile a program, it is necessary to set environment variables depending on the parallel processing MPI and the compiler. For example, if you plan to use an Intel MPI as the parallel computing MPI and an Intel compiler as the compiler, set the environment variables using the following commands shown below. Note that the Intel MPI and the Intel compiler are set as the session defaults. You can also use the following commands to set the environment variables. Parallel computing MPI Compiler Environment variable setting command Intel MPI Intel compiler module load intelmpi.intel Open MPI Intel compiler module load openmpi.intel MPICH2 Intel compiler module load mpich2.intel MPICH1 Intel compiler module load mpich.intel - Intel compiler module load intel - PGI compiler module load pgi - gcc compiler module load gcc - nnvcc compiler module load cuda-5.0 To change the environmental settings, execute the following command to delete the current settings and set the new environment variables. For example, if you change the Intel MPI and the Intel compiler to the Open MPI and the Intel compiler, execute as follows.
When using software, it is necessary to change the environment variables depending on the software package to be used. For example, use the following command to set the environment variables to use Gaussian. You can also use the following commands to set the environment variables. Software name Version Environment variable setting command Structural analysis ANSYS Multiphysics, CFX, Fluent, LS-DYNA 14.5 module load ansys14.5 ABAQUS 6.12 module load abaqus-6.12-3 Patran 2012.2 module load patran-2012.2 DEFORM-3D 10.2 module loadd deform-3d-10.2 Computational materials science PHASE (Serial version) 11.00 module load phase-11.00-serial PHASE (Parallel version) 11.00 module load phase-11.00-parallel PHASE-Viewer 3.2.0 module load phase-viewer-v320 UVSOR (Serial version) 3.42 module load uvsor-v342-serial UVSOR (Parallel version) 3.42 module load uvsor-v342-parallel OpenMX (Serial version) 3.6 module load openmx-3.6-serial OpenMX (Parallel version) 3.6 module load openmx-3.6-parallel Computational Science Gaussian 09 Rev.C.01 module load gaussian09-C.01 NWChem (Serial version) 6.1.1 module load nwchem-6.1.1-serial NWChem (Parallel version) 6.1.1 module load nwchem-6.1.1-parallel GAMESS (Serial version) 2012.R2 module load gamess-2012.r2-serial GAMESS (Parallel version) 2012.R2 module load gamess-2012.r2-parallel MPQC 3.0-alpha module load mpqc-2.4-4.10.2013.18.19 Amber, AmberTools (Serial version) 12 module load amber12-serial Amber, AmberTools (Parallel version) 12 module load amber12-parallel CONFLEX (Serial version & Parallel version) 7 module load conflex7 Technical processing MATLAB R2012a module load matlab-R2012a http://imc.tut.ac.jp/en/research/form. To cancel the software environment, execute the following command. For example, to discard the Gaussian environmental setting, use the following command.
Editing the following files automatically sets up the session environment after logging in. Example 1: To use ansys, gaussian, and conflex in your session through bash, add the following codes to ~/.bashrc. Example 2: To use OpenMPI, Intel Compiler, gaussian, and conflex in your session through csh, add the following codes to ~/.cshrc. The following command displays the currently loaded modules. The following command displays the available modules.
To compile a C/C++ program, execute the following command. To compile a FORTRAN program, execute the following command.
To compile a C/C++ program, execute the following command. To compile a FORTRAN program, execute the following command.
To compile a C/C++ program, execute the following command.
To compile a C/C++ program, execute the following command. To compile a FORTRAN program, execute the following command.
To compile a C/C++ program, execute the following command. To compile a FORTRAN program, execute the following command.
The job management system, Torque, is used to manage jobs to be executed. Always use Torque to execute jobs.
Create the Torque script and submit the job as follows using the qsub command. For example, enter the following to submit a job to rchq, a research job queue. In the script, specify the number of processors per node to request as follows. Example: To request 1 node and 16 processors per node Example: To request 4 nodes and 16 processors per node To execute a job with the specified memory capacity, add the following codes to the script. Example: To request 1 node, 16 processors per node, and 16GB memory per job To execute a job with the specified calculation node, add the following codes to the script. Example: To request csnd00 and csnd01 as the nodes and 16 processors per node * From csnd00 to csnd27 are the calculation nodes (see Hardware Configuration under System Configuration). * csnd00 and csnd01 are equipped with Tesla K20X. To execute a job on a calculation node with GPGPU, add the following codes to the script. To execute a job in a specified duration of time, add the following codes to the script. Example: To request a duration of 336 hours to execute a job The major options of the qsub command are as follows. Option Example Description -e -e filename Outputs the standard error to a specified file. When the -e option is not used, the output file is created in the directory where the qsub command was executed. The file name convention is job_name.ejob_no. -o -o filename Outputs the standard output to the specified file. When the -o option is not used, the output file is created in the directory where the qsub command was executed. The file name convention is job_name.ojob_no. -j -j join Specifies whether to merge the standard output and the standard error to a single file or not. -q -q destination Specifies the queue to submit a job. -l -l resource_list Specifies the resources required for job execution. -N -N name Specifies the job name (up to 15 characters). If a job is submitted via a script, the script file name is used as the job name by default. Otherwise, STDIN is used. -m -m mail_events Notifies the job status by an email. -M -M user_list Specifies the email address to receive the notice.
* The target queue (either eduq or rchq) to submit the job can be specified in the script.
* The target queue (either eduq or rchq) to submit the job can be specified in the script.
* When using MPICH2, always specify the -iface option. * The target queue (either eduq or rchq) to submit the job can be specified in the script.
The sample script is as follows. * vm1.dat exists in /common/ansys14.5/ansys_inc/v145/ansys/data/verif.
The sample script is as follows. Example: When using Shared Memory ANSYS Example: When using Distributed ANSYS * vm141.dat exists in /common/ansys14.5/ansys_inc/v145/ansys/data/verif
The sample script is as follows. * StaticMixer.def exists in /common/ansys14.5/ansys_inc/v145/CFX/examples.
The sample script is as follows. *StaticMixer.def exits in /common/ansys14.5/ansys_inc/v145/CFX/examples.
The sample script is as follows.
The sample script is as follows.
The sample script is as follows. * For hourglass.k, see LS-DYNA Examples (http://www.dynaexamples.com/).
The sample script is as follows. *1_mass_coarse.inp exits in /common/abaqus-6.12-3/6.12-3/samples/job_archive/samples.zip.
The sample script is as follows. * For PHASE jobs, a set of I/O files are defined as file_names.data. * Sample input files are available at /common/phase-11.00-parallel/sample/Si2/scf/.
The sample script is as follows. * For ekcal jobs, a set of I/O files are defined as file_names.data. * Sample input files are available at /common/phase-11.00-parallel/sample/Si2/band/. * (Prior to using the sample files under Si2/band/, execute the sample file in ../scf/ to create nfchgt.data.)
The sample script is as follows. * For epsmain jobs, a set of I/O files are defined as file_names.data. * Sample input files are available at /common/uvsor-v342-parallel/sample/electron/Si/eps/. * (Prior to using the sample files under Si/eps/, execute the sample file in ../scf/ to create nfchgt.data.)
The sample script is as follows. * H2O.dat exists in /common/openmx-3.6-parallel/work/. * The following line must be added to H2O.dat. * DATA.PATH /common/openmx-3.6-parallel/DFT_DATA11.
Sample input file (methane.com) e
The sample script is as follows.
The sample script is as follows.
Sample input file (h2o.nw)
The sample script is as follows.
The sample script is as follows. * exam01.inp exists in /common/gamess-2012.r2-serial/tests/standard/.
Sample input file (h2o.in)
The sample script is as follows.
Sample input file (gbin-sander) Sample input file (gbin-pmemd)
The sample script is as follows. Example: To use sander * prmtop and eq1.x exists in /common/amber12-parallel/test/4096wat. Example: To use pmemd * prmtop and eq1.x exists in /common/amber12-parallel/test/4096wat.
The sample script is as follows. Example: To use sander * prmtop and eq1.x exists in /common/amber12-parallel/test/4096wat. Example: To use pmemd * prmtop and eq1.x exists in /common/amber12-parallel/test/4096wat.
Sample input file (methane.mol)
The sample script is as follows.
Create the script and submit the job using the qsub command. The sample script is as follows.
Sample input file (matlabdemo.m)
The sample script is as follows.
The sample script is as follows.
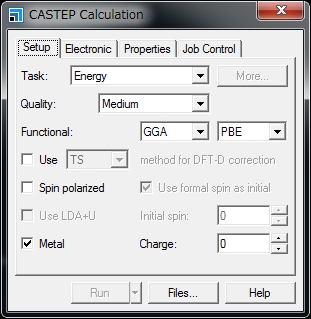
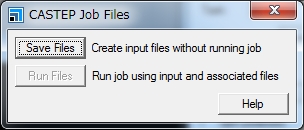
There are two ways to run Materials Studio CASTEP/DMol3 using the University’s supercomputer: (1) Submit the job from Materials Studio Visualiser to the supercomputer (only Materials Studio 2016 is supported). (2) Transfer the input file to the supercomputer using Winscp etc. and submit the job using the qsub command. This section explains about the method (2). To submit the job in the method (2), the parameters to pass the job scheduler can be changed. The following is an example using CASTEP. 1. Create an input file using Materials Studio Visualizer. In the CASTEP Calculation window, click Files…. In the CASTEP Job Files window, click Save Files. The input file is stored at: specified_path\project_name_Files\Documents\folder_created_in_above_step For example, when performing an energy calculation using acetanilide.xsd in CASTEP following the above steps, the folder is created with the name of acetanilide CASTEP Energy. The above specified_path is usually C:\Users\account_name\Documents\Materials Studio Projects. The folder name contains a blank character. Delete the blank character or replace it with underscore (_). The folder name must not contain Japanese characters. 2. Using file transfer software such as Winscp, transfer the created folder and the file it contains to the supercomputer. 3. Create a script as follows in the transferred folder and submit the job using the qsub command. * Specify the file name without an extension in the transferred folder as the third argument in RunCASTEP.sh. * Specify a value of 8 or less in =ppn. * The values for the duration of job execution and the permitted memory capacity can be changed as necessary. * Specifications in the Gateway location, Queue, and Run in parallel on fields in the Job Control tab in the CASTEP Calculation window will be ignored in job execution from the command line. The settings specified in the script used with the qsub command will be used. * The script contents vary depending on the computer and calculation modules to be used. See the following Example Scripts. 4. After the job is completed, transfer the result file to your client PC and view the results using Materials studio Visualizer. Notes on Joint Research There are eight Materials Studio licenses owned by the University for CASTEP and a further eight for Dmol3. 1One job with eight or less parallel executions can be conducted under a single license. These licenses are shared. Avoid exclusively occupying the licenses and consider other users. Example Scripts Running Materials Studio 8.0 CASTEP in the Wide-Area Coordinated Cluster System for Education and Research Running Materials Studio 8.0 DMOL3 in the Wide-Area Coordinated Cluster System for Education and Research Running Materials Studio 2016 CASTEP in the Wide-Area Coordinated Cluster System for Education and Research Running Materials Studio 2016 DMOL3 in the Wide-Area Coordinated Cluster System for Education and Research
Use the qstat command to view the job or queue status. (1) Viewing the job status (2) Viewing the queue status Major options for the qstat command are as follows. Options Usage Description -a -a Displays all jobs in the queue and jobs being executed. -Q -Q Displays the statuses of all queues in an abbreviated form. -Qf -Qf Displays the statuses of all queues in a detailed form. -q -q Displays the limitation values of the queues. -n -n Displays the calculation node name assigned to the job. -r -r Displays a list of running jobs. -i -i Displays a list of idle jobs. -u -u user_name Displays the jobs owned by the specified user. -f -f job_ID Displays details of the job that has the specified job ID.
Use the qdel command to cancel the submitted job. Check the job ID beforehand using the qstat command. Setting Up the Developmental Environment
$ module load intelmpi.intel
$ module unload environmental_settings
$ module unload intelmpi.intel
$ module load openmpi.intel
Setting Up the Software Usage Environment
$ module load gaussian09-C.01
$ module unload environmental_settings
$ module unload gaussian09-C.01
Setting Up the Session Environment
eval `modulecmd bash load ansys14.5`
eval `modulecmd bash load gaussian09-C.01`
eval `modulecmd bash load conflex7`
eval `modulecmd tcsh unload intelmpi.intel`
eval `modulecmd tcsh load openmpi.intel`
eval `modulecmd tcsh load gaussian09-C.01`
eval `modulecmd tcsh load conflex7`
$ module list
$ module avail
Compiling
Intel Compiler
$ icc source_file_name –o output_program_name
$ ifort source_file_name –o output_program_name
PGI Compiler
$ pgcc source_file_name –o output_program_name
$ pgf90 source_file_name –o output_program_name
$ pgf77 source_file_name –o output_program_name
GNU Compiler
$ gcc source_file_name –o output_program_name
Intel MPI
$ mpiicc source_file_name –o output_program_name
$ mpiifort source_file_name –o output_program_name
OpenMPI, MPICH2, MPICH1
$ mpicc source_file_name –o output_program_name
$ mpif90 source_file_name –o output_program_name
$ mpif77 source_file_name –o output_program_name
Executing Jobs
Queue Configuration
Submitting Jobs
$ qsub –q quque_name script_name
$ qsub -q rchq script_name
#PBS -l nodes=no_of_nodes:ppn=processors_per_node
#PBS -l nodes=1:ppn=16
#PBS -l nodes=4:ppn=16
#PBS -l nodes=no_of_nodes:ppn=processors_per_node,mem=memory_per_job
#PBS -l nodes=1:ppn=16,mem=16gb
#PBS -l nodes=nodeA:ppn=processors_for_nodeA+nodeB:ppn= processors_for_nodeB,・・・
#PBS -l nodes=csnd00:ppn=16+csnd01:ppn=16
#PBS -l nodes=1:GPU:ppn=1
#PBS -l walltime=hh:mm:ss
#PBS -l walltime=336:00:00
-j oe: Merges the standard error into the standard output file.
-j eo: Merges the standard output into the standard error file.
-j n: Does not merge the standard error file and the standard output file (Default).
-m n: Does not send an email.
-m a: Sends an email if the job terminates abnormally.
-m b: Sends an email when the job execution starts.
-m e: Sends an email when the job execution ends.
Sample Script: When Using an Intel MPI and an Intel Compiler
### sample
#!/bin/sh
#PBS -q rchq
#PBS -l nodes=1:ppn=16
MPI_PROCS=`wc -l $PBS_NODEFILE | awk '{print $1}'`
cd $PBS_O_WORKDIR
mpirun -np $MPI_PROCS ./a.outSample Script: When Using an OpenMPI and an Intel Compiler
### sample
#!/bin/sh
#PBS -q rchq
#PBS -l nodes=1:ppn=16
MPI_PROCS=`wc -l $PBS_NODEFILE | awk '{print $1}'`
module unload intelmpi.intel
module load openmpi.intel
cd $PBS_O_WORKDIR
mpirun -np $MPI_PROCS ./a.outSample Script: When Using an MPICH2 and an Intel Compiler
### sample
#!/bin/sh
#PBS -q rchq
#PBS -l nodes=1:ppn=16
MPI_PROCS=`wc -l $PBS_NODEFILE | awk '{print $1}'`
module unload intelmpi.intel
module load mpich2.intel
cd $PBS_O_WORKDIR
mpirun -np $MPI_PROCS -iface ib0 ./a.outSample Script: When Using an OpenMP Program
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=8
#PBS -q eduq
export OMP_NUM_THREADS=8
cd $PBS_O_WORKDIR
./a.out
Sample Script: When Using an MPI/OpenMP Hybrid Program
### sample
#!/bin/sh
#PBS -l nodes=4:ppn=8
#PBS -q eduq
export OMP_NUM_THREADS=8
cd $PBS_O_WORKDIR
sort -u $PBS_NODEFILE > hostlist
mpirun -np 4 -machinefile ./hostlist ./a.out
Sample Script: When Using a GPGPU Program
### sample
#!/bin/sh
#PBS -l nodes=1:GPU:ppn=1
#PBS -q eduq
cd $PBS_O_WORKDIR
module load cuda-5.0
./a.out
ANSYS Multiphysics Job
Single Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load ansys14.5
cd $PBS_O_WORKDIR
ansys145 -b nolist -p AA_T_A -i vm1.dat -o vm1.out -j vm1
Parallel Jobs
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=4
#PBS -q eduq
module load ansys14.5
cd $PBS_O_WORKDIR
ansys145 -b nolist -p AA_T_A -i vm141.dat -o vm141.out -j vm141 -np 4
### sample
#!/bin/sh
#PBS -l nodes=2:ppn=2
#PBS -q eduq
module load ansys14.5
cd $PBS_O_WORKDIR
ansys145 -b nolist -p AA_T_A -i vm141.dat -o vm141.out -j vm141 -np 4 -dis
ANSYS CFX Job
Single Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load ansys14.5
cd $PBS_O_WORKDIR
cfx5solve -def StaticMixer.def
Parallel Jobs
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=4
#PBS -q eduq
module load ansys14.5
cd $PBS_O_WORKDIR
cfx5solve -def StaticMixer.def -part 4 -start-method 'Intel MPI Local Parallel'
ANSYS Fluent Job
Single Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load ansys14.5
cd $PBS_O_WORKDIR
fluent 3ddp -g -i input-3d > stdout.txt 2>&1
Parallel Jobs
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=4
#PBS -q eduq
module load ansys14.5
cd $PBS_O_WORKDIR
fluent 3ddp -g -i input-3d -t4 -mpi=intel > stdout.txt 2>&1
ANSYS LS-DYNA Job
Single Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load ansys14.5
cd $PBS_O_WORKDIR
lsdyna145 i=hourglass.k memory=100m
ABAQUS Job
Single Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load abaqus-6.12-3
cd $PBS_O_WORKDIR
abaqus job=1_mass_coarse
PHASE Job
Parallel Jobs (phase command: SCF calculation)
### sample
#!/bin/sh
#PBS -q eduq
#PBS -l nodes=2:ppn=2
cd $PBS_O_WORKDIR
module load phase-11.00-parallel
mpirun -np 4 phase
Single Job (ekcal command: Band calculation)
### sample
#!/bin/sh
#PBS -q eduq
#PBS -l nodes=1:ppn=1
cd $PBS_O_WORKDIR
module load phase-11.00-parallel
ekcal
UVSOR Job
Parallel Jobs (epsmain command: Dielectric constant calculation)
### sample
#!/bin/sh
#PBS -q eduq
#PBS -l nodes=2:ppn=2
cd $PBS_O_WORKDIR
module load uvsor-v342-parallel
mpirun -np 4 epsmain
OpenMX Job
Parallel Jobs
### sample
#!/bin/bash
#PBS -l nodes=2:ppn=2
cd $PBS_O_WORKDIR
module load openmx-3.6-parallel
export LD_LIBRARY_PATH=/common/intel-2013/composer_xe_2013.1.117/mkl/lib/intel64:$LD_LIBRARY_PATH
mpirun -4 openmx H2O.dat
GAUSSIAN Job
%NoSave
%Mem=512MB
%NProcShared=4
%chk=methane.chk
#MP2/6-31G opt
methane
0 1
C -0.01350511 0.30137653 0.27071342
H 0.34314932 -0.70743347 0.27071342
H 0.34316773 0.80577472 1.14436492
H 0.34316773 0.80577472 -0.60293809
H -1.08350511 0.30138971 0.27071342
Single Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load gaussian09-C.01
cd $PBS_O_WORKDIR
g09 methane.com
Parallel Jobs
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=4,mem=3gb,pvmem=3gb
#PBS -q eduq
cd $PBS_O_WORKDIR
module load gaussian09-C.01
g09 methane.com
NWChem Job
echo
start h2o
title h2o
geometry units au
O 0 0 0
H 0 1.430 -1.107
H 0 -1.430 -1.107
end
basis
* library 6-31g**
end
scf
direct; print schwarz; profile
end
task scf
Parallel Jobs
### sample
#!/bin/sh
#PBS -q eduq
#PBS -l nodes=2:ppn=2
export NWCHEM_TOP=/common/nwchem-6.1.1-parallel
cd $PBS_O_WORKDIR
module load nwchem.parallel
mpirun -np 4 nwchem h2o.nw
GAMESS Job
Single Job
### sample
#!/bin/bash
#PBS -q eduq
#PBS -l nodes=1:ppn=1
module load gamess-2012.r2-serial
mkdir -p /work/$USER/scratch/$PBS_JOBID
mkdir -p $HOME/scr
rm -f $HOME/scr/exam01.dat
rungms exam01.inp 00 1
MPQC Job
% HF/STO-3G SCF water
method: HF
basis: STO-3G
molecule:
O 0.172 0.000 0.000
H 0.745 0.000 0.754
H 0.745 0.000 -0.754Parallel Jobs
### sample
#!/bin/sh
#PBS -q rchq
#PBS -l nodes=2:ppn=2
MPQCDIR=/common/mpqc-2.4-4.10.2013.18.19
cd $PBS_O_WORKDIR
module load mpqc-2.4-4.10.2013.18.19
mpirun -np 4 mpqc -o h2o.out h2o.in
AMBER Job
short md, nve ensemble
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2, tol=0.0000001,
nstlim=100, ntt=0,
ntpr=1, ntwr=10000,
dt=0.001,
/
&ewald
nfft1=50, nfft2=50, nfft3=50, column_fft=1,
/
EOF
short md, nve ensemble
&cntrl
ntx=5, irest=1,
ntc=2, ntf=2, tol=0.0000001,
nstlim=100, ntt=0,
ntpr=1, ntwr=10000,
dt=0.001,
/
&ewald
nfft1=50, nfft2=50, nfft3=50,
/
EOF
Single Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load amber12-serial
cd $PBS_O_WORKDIR
sander -i gbin-sander -p prmtop -c eq1.x
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load amber12-serial
cd $PBS_O_WORKDIR
pmemd -i gbin-pmemd -p prmtop -c eq1.x
Parallel Jobs
### sample
#!/bin/sh
#PBS -l nodes=2:ppn=4
#PBS -q eduq
module unload intelmpi.intel
module load openmpi.intel
module load amber12-parallel
cd $PBS_O_WORKDIR
mpirun -np 8 sander.MPI -i gbin-sander -p prmtop -c eq1.x
### sample
#!/bin/sh
#PBS -l nodes=2:ppn=4
#PBS -q eduq
module unload intelmpi.intel
module load openmpi.intel
module load amber12-parallel
cd $PBS_O_WORKDIR
mpirun -np 8 pmemd.MPI -i gbin-pmemd -p prmtop -c eq1.x
CONFLEX Job
Sample
Chem3D Core 13.0.009171314173D
5 4 0 0 0 0 0 0 0 0999 V2000
0.1655 0.7586 0.0176 H 0 0 0 0 0 0 0 0 0 0 0 0
0.2324 -0.3315 0.0062 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7729 -0.7586 0.0063 H 0 0 0 0 0 0 0 0 0 0 0 0
0.7729 -0.6729 0.8919 H 0 0 0 0 0 0 0 0 0 0 0 0
0.7648 -0.6523 -0.8919 H 0 0 0 0 0 0 0 0 0 0 0 0
2 1 1 0
2 3 1 0
2 4 1 0
2 5 1 0
M ENDSingle Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=1
#PBS -q eduq
c
cd $PBS_O_WORKDIR
flex7a1.ifc12.Linux.exe -par /common/conflex7/par methane
Parallel Jobs
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=8
#PBS -q eduq
module load conflex7
cd $PBS_O_WORKDIR
mpirun -np 8 flex7a1.ifc12.iMPI.Linux.exe -par /common/conflex7/par methane
MATLAB Job
% Sample
n=6000;X=rand(n,n);Y=rand(n,n);tic; Z=X*Y;toc
% after finishing work
switch getenv('PBS_ENVIRONMENT')
case {'PBS_INTERACTIVE',''}
disp('Job finished. Results not yet saved.');
case 'PBS_BATCH'
disp('Job finished. Saving results.')
matfile=sprintf('matlab-%s.mat',getenv('PBS_JOBID'));
save(matfile);
exit
otherwise
disp([ 'Unknown PBS environment ' getenv('PBS_ENVIRONMENT')]);
endSingle Job
### sample
#!/bin/bash
#PBS -l nodes=1:ppn=1
#PBS -q eduq
module load matlab-R2012a
cd $PBS_O_WORKDIR
matlab -nodisplay -nodesktop -nosplash -nojvm -r "maxNumCompThreads(1); matlabdemo"
Parallel Jobs
### sample
#!/bin/bash
#PBS -l nodes=1:ppn=4
#PBS -q eduq
module load matlab-R2012a
cd $PBS_O_WORKDIR
matlab -nodisplay -nodesktop -nosplash -nojvm -r "maxNumCompThreads(4); matlabdemo"
CASTEP/DMol3 Job
### sample
#!/bin/sh
#PBS -l nodes=1:ppn=8,mem=24gb,pmem=3gb,vmem=24gb,pvmem=3gb
#PBS -l walltime=24:00:00
#PBS -q rchq
MPI_PROCS=`wc -l $PBS_NODEFILE | awk '{print $1}'`
cd $PBS_O_WORKDIR
cp /common/accelrys/cdev0/MaterialsStudio8.0/etc/CASTEP/bin/RunCASTEP.sh .
perl -i -pe 's/\r//' *
./RunCASTEP.sh -np $MPI_PROCS acetanilide### sample
#!/bin/sh
#PBS -l nodes=1:ppn=8,mem=50000mb,pmem=6250mb,vmem=50000mb,pvmem=6250mb
#PBS -l walltime=24:00:00
#PBS -q wLrchq
MPI_PROCS=`wc -l $PBS_NODEFILE | awk '{print $1}'`
cd $PBS_O_WORKDIR
cp /common/accelrys/wdev0/MaterialsStudio8.0/etc/CASTEP/bin/RunCASTEP.sh .
perl -i -pe 's/\r//' *
./RunCASTEP.sh -np $MPI_PROCS acetanilide### sample
#!/bin/sh
#PBS -l nodes=1:ppn=8,mem=50000mb,pmem=6250mb,vmem=50000mb,pvmem=6250mb
#PBS -l walltime=24:00:00
#PBS -q wLrchq
MPI_PROCS=`wc -l $PBS_NODEFILE | awk '{print $1}'`
cd $PBS_O_WORKDIR
cp /common/accelrys/wdev0/MaterialsStudio8.0/etc/DMol3/bin/RunDMol3.sh .
perl -i -pe 's/\r//' *
./RunDMol3.sh -np $MPI_PROCS acetanilide### sample
#!/bin/sh
#PBS -l nodes=1:ppn=8,mem=50000mb,pmem=6250mb,vmem=50000mb,pvmem=6250mb
#PBS -l walltime=24:00:00
#PBS -q wLrchq
MPI_PROCS=`wc -l $PBS_NODEFILE | awk '{print $1}'`
cd $PBS_O_WORKDIR
cp /common/accelrys/wdev0/MaterialsStudio2016/etc/CASTEP/bin/RunCASTEP.sh .
perl -i -pe 's/\r//' *
./RunCASTEP.sh -np $MPI_PROCS acetanilide### sample
#!/bin/sh
#PBS -l nodes=1:ppn=8,mem=50000mb,pmem=6250mb,vmem=50000mb,pvmem=6250mb
#PBS -l walltime=24:00:00
#PBS -q wLrchq
MPI_PROCS=`wc -l $PBS_NODEFILE | awk '{print $1}'`
cd $PBS_O_WORKDIR
cp /common/accelrys/wdev0/MaterialsStudio2016/etc/DMol3/bin/RunDMol3.sh .
perl -i -pe 's/\r//' *
./RunDMol3.sh -np $MPI_PROCS acetanilideViewing Job Status
qstat -a
qstat -Q
Canceling a Submitted Job
qdel jobID